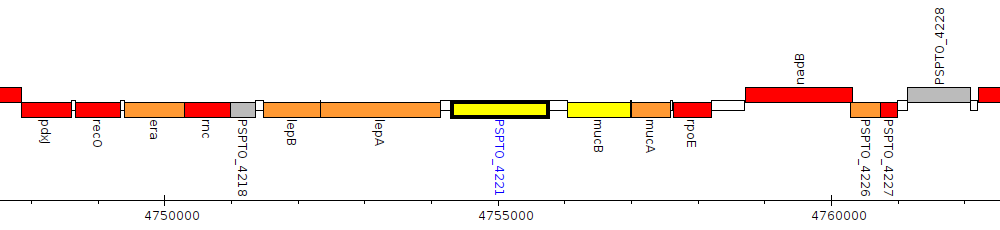
Pseudomonas syringae pv. tomato DC3000, PSPTO_4221
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0004252 | serine-type endopeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR00834
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006508 | proteolysis |
Inferred from Sequence Model
Term mapped from: InterPro:PR00834
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005515 | protein binding |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:2.30.42.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pst02020 | Two-component system | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pst01503 | Cationic antimicrobial peptide (CAMP) resistance | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF50156 | PDZ domain-like | IPR036034 | PDZ superfamily | 246 | 371 | 1.01E-23 |
| SUPERFAMILY | SSF50156 | PDZ domain-like | IPR036034 | PDZ superfamily | 390 | 477 | 1.58E-13 |
| Pfam | PF13180 | PDZ domain | IPR001478 | PDZ domain | 277 | 366 | 1.2E-16 |
| CDD | cd00987 | PDZ_serine_protease | - | - | 275 | 364 | 2.359E-26 |
| Gene3D | G3DSA:2.30.42.10 | - | IPR036034 | PDZ superfamily | 386 | 479 | 3.8E-17 |
| Gene3D | G3DSA:2.30.42.10 | - | IPR036034 | PDZ superfamily | 273 | 371 | 1.1E-28 |
| Gene3D | G3DSA:2.40.10.120 | - | - | - | 30 | 268 | 6.7E-85 |
| SMART | SM00228 | pdz_new | IPR001478 | PDZ domain | 389 | 468 | 7.5E-9 |
| SUPERFAMILY | SSF50494 | Trypsin-like serine proteases | IPR009003 | Peptidase S1, PA clan | 6 | 284 | 5.89E-70 |
| PRINTS | PR00834 | HtrA/DegQ protease family signature | IPR001940 | Peptidase S1C | 229 | 246 | 1.5E-52 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 79 | 99 | - |
| SMART | SM00228 | pdz_new | IPR001478 | PDZ domain | 275 | 357 | 2.0E-14 |
| Pfam | PF13180 | PDZ domain | IPR001478 | PDZ domain | 391 | 470 | 4.0E-9 |
| PRINTS | PR00834 | HtrA/DegQ protease family signature | IPR001940 | Peptidase S1C | 173 | 197 | 1.5E-52 |
| FunFam | G3DSA:2.40.10.120:FF:000007 | Periplasmic serine endoprotease DegP-like | - | - | 31 | 268 | 3.2E-99 |
| NCBIfam | TIGR02037 | JCVI: Do family serine endopeptidase | IPR011782 | Peptidase S1C, Do | 32 | 475 | 0.0 |
| PANTHER | PTHR22939 | SERINE PROTEASE FAMILY S1C HTRA-RELATED | - | - | 397 | 474 | 8.6E-97 |
| PRINTS | PR00834 | HtrA/DegQ protease family signature | IPR001940 | Peptidase S1C | 112 | 124 | 1.5E-52 |
| PRINTS | PR00834 | HtrA/DegQ protease family signature | IPR001940 | Peptidase S1C | 315 | 327 | 1.5E-52 |
| Pfam | PF13365 | Trypsin-like peptidase domain | - | - | 103 | 236 | 2.8E-34 |
| CDD | cd00987 | PDZ_serine_protease | - | - | 390 | 475 | 2.26428E-15 |
| Pfam | PF02163 | Peptidase family M50 | IPR008915 | Peptidase M50 | 244 | 375 | 8.5E-10 |
| PRINTS | PR00834 | HtrA/DegQ protease family signature | IPR001940 | Peptidase S1C | 207 | 224 | 1.5E-52 |
| PRINTS | PR00834 | HtrA/DegQ protease family signature | IPR001940 | Peptidase S1C | 133 | 153 | 1.5E-52 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.