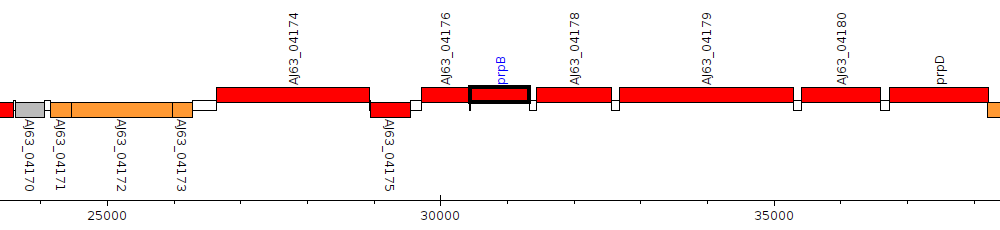
Pseudomonas aeruginosa 3576, AJ63_04177 (prpB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0046421 | methylisocitrate lyase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR02317
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0019629 | propionate catabolic process, 2-methylcitrate cycle |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR02317
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF51621
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.20.20.60 | - | IPR040442 | Pyruvate kinase-like domain superfamily | 2 | 297 | 6.1E-105 |
| SUPERFAMILY | SSF51621 | Phosphoenolpyruvate/pyruvate domain | IPR015813 | Pyruvate/Phosphoenolpyruvate kinase-like domain superfamily | 8 | 292 | 2.02E-100 |
| PANTHER | PTHR42905 | PHOSPHOENOLPYRUVATE CARBOXYLASE | - | - | 4 | 287 | 1.3E-84 |
| Coils | Coil | Coil | - | - | 281 | 298 | - |
| Pfam | PF13714 | Phosphoenolpyruvate phosphomutase | - | - | 19 | 256 | 8.3E-46 |
| CDD | cd00377 | ICL_PEPM | IPR039556 | ICL/PEPM domain | 12 | 255 | 1.40963E-94 |
| NCBIfam | TIGR02317 | JCVI: methylisocitrate lyase | IPR012695 | 2-methylisocitrate lyase | 9 | 293 | 0.0 |
| FunFam | G3DSA:3.20.20.60:FF:000009 | 2-methylisocitrate lyase | - | - | 4 | 297 | 0.0 |
| Hamap | MF_01939 | 2-methylisocitrate lyase [prpB]. | IPR012695 | 2-methylisocitrate lyase | 4 | 295 | 51.604546 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.