In order to generate a more inclusive dataset of Pseudomonas genes mapped to putative in-paralogs and putative orthologs in other Pseudomonas species/strains, we developed a Pseudomonas Orthologous Groups classification system.
To generate ortholog groups, pair-wise DIAMOND searches were run on all genomes in the database to find reciprocal best hits (RBHs) for each gene. These analyses often resulted in multiple candidate genes for RBH status, which were narrowed down by examining the similarity between the query's flanking genes and the hit's flanking genes. If two candidate genes were directly adjacent, they where both accepted as RBHs that involve putative in-parology.
Pairwise intra-genome DIAMOND searches were also performed to acquire in-paralog information (i.e. gene duplications occurring after species divergence). If two genes in one genome were reciprocally more similar to each other than to any gene in the other genomes, the two genes were designated putative in-paralogs. Ortholog groups are built by starting with a seed gene and then adding all genes to which there is a RBH or in-paralog relationship.
Every new gene added to an ortholog group was then treated as a seed gene and the addition process was repeated until all qualifying genes had been added. The result was the development of orthologous groups, specifically generated for Pseudomonas species genomes, which can be used to sort search results.
Pseudomonas Ortholog Group POG000034
| Strain | Locus Tag | Description | Same-Strain Members | Fragment ? | |
|---|---|---|---|---|---|
| Pseudomonas sp. URIL14HWK12:I6 | H043_RS0111905 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas sp. URIL14HWK12:I7 | H032_RS0108350 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas sp. URMO17WK12:I11 - Assembly GCF_000514235.1 | H041_RS0121310 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas sp. URMO17WK12:I12 | D882_RS0108565 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas sp. URMO17WK12:I8 | F639_RS0113450 |
LuxR family transcriptional regulator
|
2 same-strain members: F639_RS0113450 F639_RS0116095 |
|
|
| Pseudomonas sp. URMO17WK12:I8 | F639_RS0116095 |
LuxR family transcriptional regulator
|
2 same-strain members: F639_RS0113450 F639_RS0116095 |
|
|
| Pseudomonas sp. VLB120 | PVLB_07720 |
two component LuxR family transcriptional regulator
|
2 same-strain members: PVLB_05445 PVLB_07720 |
|
|
| Pseudomonas sp. VLB120 | PVLB_05445 |
two component LuxR family transcriptional regulator
|
2 same-strain members: PVLB_05445 PVLB_07720 |
|
|
| Pseudomonas sp. WCS358 | PC358_RS13145 |
LuxR family transcriptional regulator
|
2 same-strain members: PC358_RS13145 PC358_RS22410 |
|
|
| Pseudomonas sp. WCS358 | PC358_RS22410 |
LuxR family transcriptional regulator
|
2 same-strain members: PC358_RS13145 PC358_RS22410 |
|
|
| Pseudomonas stutzeri KOS6 | B597_020110 |
response regulator
|
1 member |
|
|
| Pseudomonas stutzeri RCH2 | Psest_2705 |
response regulator containing a CheY-like receiver domain and an HTH DNA-binding domain
|
1 member |
|
|
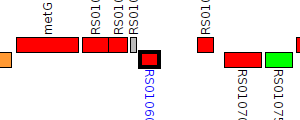
| Pseudomonas syringae B576 | NG81_RS01060 |
LuxR family transcriptional regulator
|
1 member |
|
|
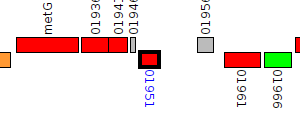
| Pseudomonas syringae BRIP39023 | A988_01951 |
LuxR family transcriptional regulator
|
1 member |
|
|
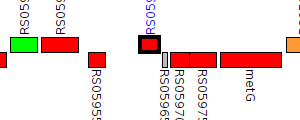
| Pseudomonas syringae CC1557 | N018_RS05960 |
LuxR family transcriptional regulator
|
1 member |
|
|
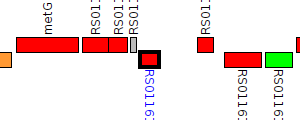
| Pseudomonas syringae DSM 10604 | PSSR_RS0116160 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas syringae pv. actinidiae ICMP 9617 | A250_RS06385 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas syringae pv. coryli NCPPB 4273 | P406_RS24645 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas syringae pv. syringae 41a | PSS41A_RS07350 |
LuxR family transcriptional regulator
|
1 member |
|
|
| Pseudomonas syringae pv. syringae B301D | PSYRB_RS19725 |
LuxR family transcriptional regulator
|
1 member |
|