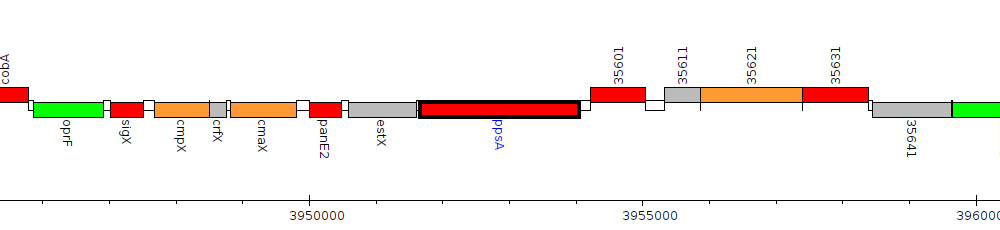
Pseudomonas aeruginosa LESB58, PALES_35591 (ppsA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0016310 | phosphorylation |
Inferred from Sequence Model
Term mapped from: InterPro:PF02896
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006090 | pyruvate metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR43030
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008986 | pyruvate, water dikinase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR43030
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016301 | kinase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF01326
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016772 | transferase activity, transferring phosphorus-containing groups |
Inferred from Sequence Model
Term mapped from: InterPro:PF02896
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF01326
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF51621
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Pyruvate metabolism |
ECO:0000037
not_recorded |
|||
| PseudoCAP | Reductive carboxylate cycle (CO2 fixation) |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| NCBIfam | TIGR01418 | JCVI: pyruvate, water dikinase | IPR006319 | Phosphoenolpyruvate synthase | 5 | 786 | 0.0 |
| Gene3D | G3DSA:3.50.30.10 | Phosphohistidine domain | - | - | 358 | 476 | 9.8E-35 |
| Gene3D | G3DSA:3.30.1490.20 | - | IPR013815 | ATP-grasp fold, subdomain 1 | 2 | 192 | 9.2E-67 |
| Pfam | PF01326 | Pyruvate phosphate dikinase, AMP/ATP-binding domain | IPR002192 | Pyruvate phosphate dikinase, AMP/ATP-binding | 17 | 342 | 1.9E-119 |
| SUPERFAMILY | SSF56059 | Glutathione synthetase ATP-binding domain-like | - | - | 1 | 384 | 9.3E-109 |
| PRINTS | PR01736 | Phosphoenolpyruvate-protein phosphotransferase signature | - | - | 710 | 725 | 7.8E-6 |
| PIRSF | PIRSF000854 | PEP_synthase | IPR006319 | Phosphoenolpyruvate synthase | 1 | 791 | 0.0 |
| SUPERFAMILY | SSF52009 | Phosphohistidine domain | IPR036637 | Phosphohistidine domain superfamily | 346 | 463 | 1.83E-33 |
| SUPERFAMILY | SSF51621 | Phosphoenolpyruvate/pyruvate domain | IPR015813 | Pyruvate/Phosphoenolpyruvate kinase-like domain superfamily | 461 | 786 | 1.05E-99 |
| Pfam | PF02896 | PEP-utilising enzyme, PEP-binding domain | IPR000121 | PEP-utilising enzyme, C-terminal | 481 | 780 | 1.3E-62 |
| FunFam | G3DSA:3.20.20.60:FF:000010 | Phosphoenolpyruvate synthase | - | - | 477 | 788 | 0.0 |
| PRINTS | PR01736 | Phosphoenolpyruvate-protein phosphotransferase signature | - | - | 693 | 708 | 7.8E-6 |
| Pfam | PF00391 | PEP-utilising enzyme, mobile domain | IPR008279 | PEP-utilising enzyme, mobile domain | 383 | 454 | 1.6E-25 |
| FunFam | G3DSA:3.30.470.20:FF:000017 | Phosphoenolpyruvate synthase | - | - | 193 | 356 | 7.2E-87 |
| FunFam | G3DSA:3.50.30.10:FF:000002 | Phosphoenolpyruvate synthase | - | - | 358 | 476 | 8.4E-67 |
| FunFam | G3DSA:3.30.1490.20:FF:000010 | Phosphoenolpyruvate synthase | - | - | 1 | 192 | 7.4E-97 |
| PRINTS | PR01736 | Phosphoenolpyruvate-protein phosphotransferase signature | - | - | 746 | 758 | 7.8E-6 |
| PANTHER | PTHR43030 | PHOSPHOENOLPYRUVATE SYNTHASE | IPR006319 | Phosphoenolpyruvate synthase | 3 | 788 | 0.0 |
| Gene3D | G3DSA:3.20.20.60 | - | IPR040442 | Pyruvate kinase-like domain superfamily | 477 | 787 | 1.6E-102 |
| Gene3D | G3DSA:3.30.470.20 | - | - | - | 193 | 356 | 7.3E-57 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.