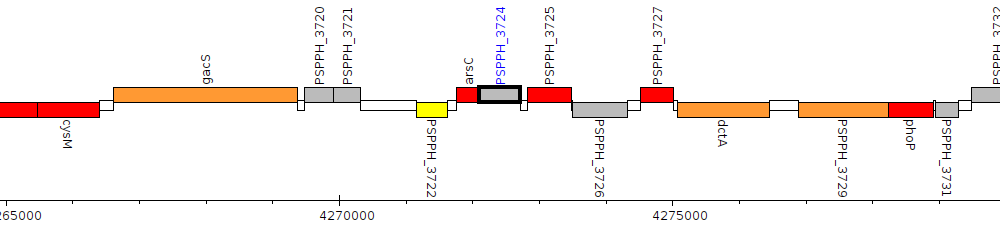
Pseudomonas savastanoi pv. phaseolicola 1448A, PSPPH_3724
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF03358
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0010181 | FMN binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01755
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01755
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | psp01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | psp00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.50.360 | - | IPR029039 | Flavoprotein-like superfamily | 1 | 197 | 2.3E-66 |
| FunFam | G3DSA:3.40.50.360:FF:000001 | NAD(P)H dehydrogenase (Quinone) FQR1-like | - | - | 1 | 198 | 6.1E-66 |
| SUPERFAMILY | SSF52218 | Flavoproteins | IPR029039 | Flavoprotein-like superfamily | 5 | 196 | 2.23E-61 |
| PANTHER | PTHR30546 | FLAVODOXIN-RELATED PROTEIN WRBA-RELATED | - | - | 6 | 196 | 3.6E-40 |
| NCBIfam | TIGR01755 | JCVI: NAD(P)H:quinone oxidoreductase, type IV | IPR010089 | Flavoprotein WrbA-like | 6 | 195 | 1.4E-43 |
| Pfam | PF03358 | NADPH-dependent FMN reductase | IPR005025 | NADPH-dependent FMN reductase-like | 12 | 141 | 6.5E-9 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.