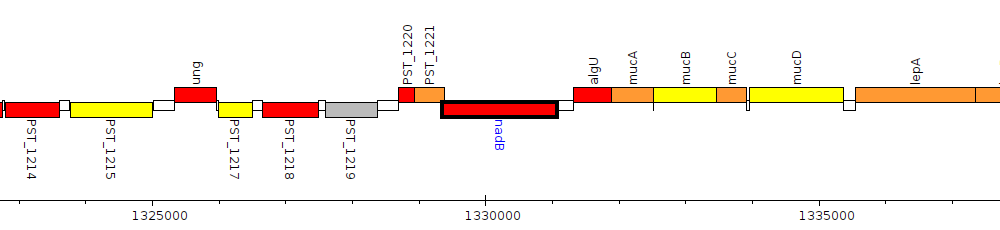
Pseudomonas stutzeri A1501, PST_1222 (nadB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009435 | NAD biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00551
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF46977
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008734 | L-aspartate oxidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00551
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | psa00760 | Nicotinate and nicotinamide metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | psa01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | psa00250 | Alanine, aspartate and glutamate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.50.50.60 | - | IPR036188 | FAD/NAD(P)-binding domain superfamily | 46 | 442 | 0.0 |
| Pfam | PF02910 | Fumarate reductase flavoprotein C-term | IPR015939 | Fumarate reductase/succinate dehydrogenase flavoprotein-like, C-terminal | 479 | 559 | 6.1E-12 |
| NCBIfam | TIGR00551 | JCVI: L-aspartate oxidase | IPR005288 | L-aspartate oxidase | 45 | 558 | 0.0 |
| FunFam | G3DSA:1.20.58.100:FF:000002 | L-aspartate oxidase | - | - | 464 | 573 | 8.2E-47 |
| SUPERFAMILY | SSF56425 | Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain | IPR027477 | Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain superfamily | 276 | 391 | 4.93E-43 |
| PIRSF | PIRSF000171 | SDHA_APRA_LASPO | - | - | 35 | 570 | 7.7E-34 |
| SUPERFAMILY | SSF51905 | FAD/NAD(P)-binding domain | IPR036188 | FAD/NAD(P)-binding domain superfamily | 36 | 486 | 6.22E-73 |
| Gene3D | G3DSA:3.90.700.10 | Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain | IPR027477 | Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain superfamily | 286 | 399 | 0.0 |
| SUPERFAMILY | SSF46977 | Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain | IPR037099 | Fumarate reductase/succinate dehydrogenase flavoprotein-like, C-terminal domain superfamily | 463 | 565 | 2.88E-30 |
| FunFam | G3DSA:3.90.700.10:FF:000002 | L-aspartate oxidase | - | - | 286 | 399 | 8.0E-53 |
| Pfam | PF00890 | FAD binding domain | IPR003953 | FAD-dependent oxidoreductase 2, FAD binding domain | 46 | 430 | 3.7E-102 |
| PRINTS | PR00368 | FAD-dependent pyridine nucleotide reductase signature | - | - | 47 | 66 | 8.0E-9 |
| PRINTS | PR00368 | FAD-dependent pyridine nucleotide reductase signature | - | - | 393 | 415 | 8.0E-9 |
| FunFam | G3DSA:3.50.50.60:FF:000060 | L-aspartate oxidase | - | - | 46 | 299 | 3.0E-112 |
| PANTHER | PTHR42716 | L-ASPARTATE OXIDASE | IPR005288 | L-aspartate oxidase | 35 | 564 | 0.0 |
| Gene3D | G3DSA:1.20.58.100 | - | - | - | 463 | 575 | 3.8E-46 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.