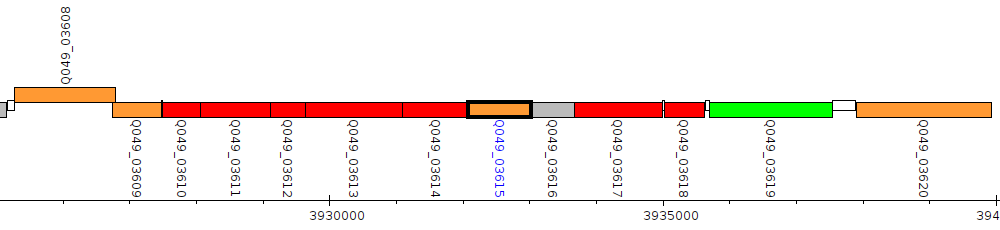
Pseudomonas aeruginosa BWHPSA044, Q049_03615
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009236 | cobalamin biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00024
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00024
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0048472 | threonine-phosphate decarboxylase activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00024
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF03186 | CobD/Cbib protein | IPR004485 | Cobalamin biosynthesis CobD/CbiB | 7 | 290 | 3.9E-95 |
| Hamap | MF_00024 | Cobalamin biosynthesis protein CbiB [cbiB]. | IPR004485 | Cobalamin biosynthesis CobD/CbiB | 1 | 312 | 108.250122 |
| NCBIfam | TIGR00380 | JCVI: adenosylcobinamide-phosphate synthase CbiB | IPR004485 | Cobalamin biosynthesis CobD/CbiB | 8 | 305 | 3.2E-57 |
| PANTHER | PTHR34308 | COBALAMIN BIOSYNTHESIS PROTEIN CBIB | IPR004485 | Cobalamin biosynthesis CobD/CbiB | 2 | 310 | 2.3E-106 |
| Pfam | PF17113 | Regulatory signalling modulator protein AmpE | IPR031347 | Protein AmpE | 156 | 229 | 3.7E-6 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.