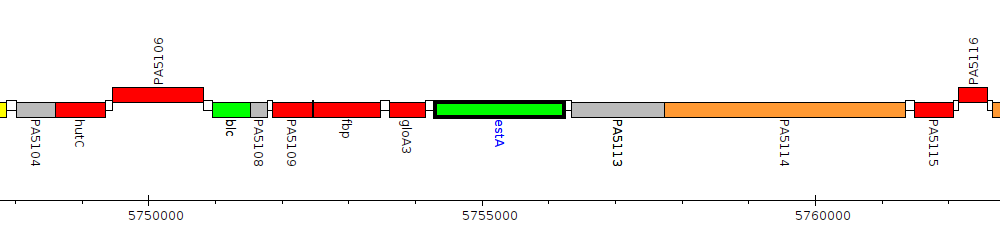
Pseudomonas aeruginosa PAO1, PA5112 (estA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0071973 | bacterial-type flagellum-dependent cell motility | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
17631636 | Reviewed by curator |
| Molecular Function | GO:0052689 | carboxylic ester hydrolase activity | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
17631636 | Reviewed by curator |
| Biological Process | GO:0044010 | single-species biofilm formation | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
17631636 | Reviewed by curator |
| Biological Process | GO:0009247 | glycolipid biosynthetic process | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
17631636 | Reviewed by curator |
| Biological Process | GO:0071978 | bacterial-type flagellum-dependent swarming motility | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
20360178 | Reviewed by curator |
| Molecular Function | GO:0016788 | hydrolase activity, acting on ester bonds |
Inferred from Sequence Model
Term mapped from: InterPro:PF00657
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Type V secretion system |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SMART | SM00869 | Autotransporter_2 | IPR005546 | Autotransporter beta-domain | 370 | 636 | 1.9E-36 |
| PANTHER | PTHR45642 | GDSL ESTERASE/LIPASE EXL3 | - | - | 29 | 325 | 1.6E-16 |
| FunFam | G3DSA:2.40.128.130:FF:000004 | Autotransporter domain-containing esterase | - | - | 339 | 646 | 0.0 |
| SUPERFAMILY | SSF103515 | Autotransporter | IPR036709 | Autotransporter beta-domain superfamily | 349 | 646 | 5.36E-47 |
| Pfam | PF00657 | GDSL-like Lipase/Acylhydrolase | IPR001087 | GDSL lipase/esterase | 32 | 325 | 1.2E-17 |
| CDD | cd01847 | Triacylglycerol_lipase_like | - | - | 29 | 330 | 2.41574E-58 |
| Pfam | PF03797 | Autotransporter beta-domain | IPR005546 | Autotransporter beta-domain | 374 | 626 | 5.5E-48 |
| FunFam | G3DSA:3.40.50.1110:FF:000027 | Esterase EstA | - | - | 19 | 338 | 0.0 |
| Gene3D | G3DSA:3.40.50.1110 | SGNH hydrolase | IPR036514 | SGNH hydrolase superfamily | 19 | 338 | 6.7E-94 |
| Gene3D | G3DSA:2.40.128.130 | - | IPR036709 | Autotransporter beta-domain superfamily | 339 | 646 | 4.8E-84 |
| PIRSF | PIRSF037375 | EstA | IPR017186 | Lipase, autotransporter EstA | 2 | 646 | 0.0 |
| NCBIfam | NF041609 | NCBIFAM: esterase EstP | IPR048099 | Esterase EstA/EstP | 4 | 646 | 0.0 |
| SUPERFAMILY | SSF52266 | SGNH hydrolase | - | - | 30 | 330 | 1.53E-15 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.