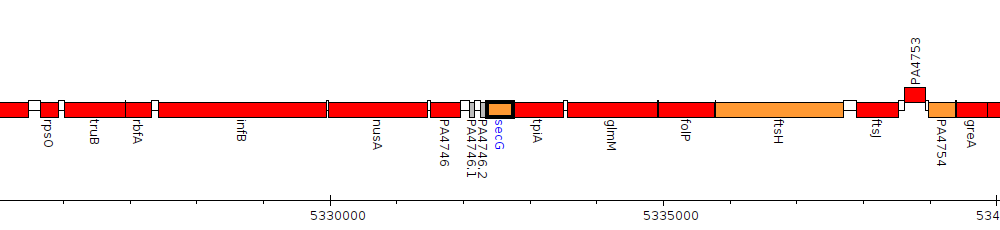
Pseudomonas aeruginosa PAO1, PA4747 (secG)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0065002 | intracellular protein transmembrane transport | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
7650029 | Reviewed by curator |
| Biological Process | GO:0043952 | protein transport by the Sec complex | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
7650029 | Reviewed by curator |
| Biological Process | GO:0006886 | intracellular protein transport | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
7650029 | Reviewed by curator |
| Molecular Function | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR01651
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009306 | protein secretion |
Inferred from Sequence Model
Term mapped from: InterPro:PR01651
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:PR01651
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae03070 | Bacterial secretion system | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae02024 | Quorum sensing | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae03060 | Protein export | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR01651 | Protein-export SecG membrane protein signature | IPR004692 | Preprotein translocase SecG subunit | 39 | 55 | 7.0E-41 |
| PANTHER | PTHR34182 | PROTEIN-EXPORT MEMBRANE PROTEIN SECG | IPR004692 | Preprotein translocase SecG subunit | 3 | 111 | 3.5E-29 |
| PRINTS | PR01651 | Protein-export SecG membrane protein signature | IPR004692 | Preprotein translocase SecG subunit | 4 | 24 | 7.0E-41 |
| Pfam | PF03840 | Preprotein translocase SecG subunit | IPR004692 | Preprotein translocase SecG subunit | 5 | 73 | 2.1E-19 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 93 | 129 | - |
| PRINTS | PR01651 | Protein-export SecG membrane protein signature | IPR004692 | Preprotein translocase SecG subunit | 24 | 38 | 7.0E-41 |
| PRINTS | PR01651 | Protein-export SecG membrane protein signature | IPR004692 | Preprotein translocase SecG subunit | 55 | 77 | 7.0E-41 |
| NCBIfam | TIGR00810 | JCVI: preprotein translocase subunit SecG | IPR004692 | Preprotein translocase SecG subunit | 5 | 74 | 4.0E-24 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.