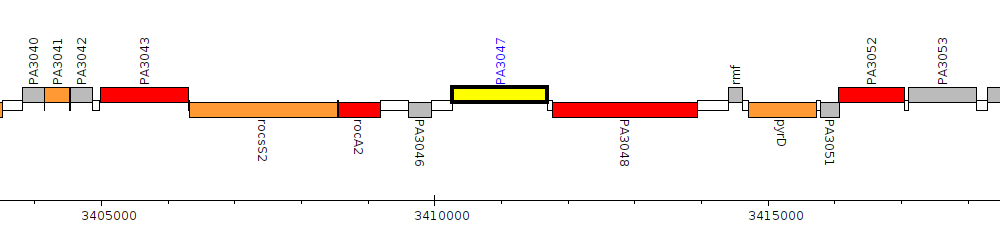
Pseudomonas aeruginosa PAO1, PA3047
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Cellular Component | GO:0042597 | periplasmic space | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
18266856 | Reviewed by curator |
| Molecular Function | GO:0004175 | endopeptidase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
25495032 | Reviewed by curator |
| Molecular Function | GO:0004185 | serine-type carboxypeptidase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
25495032 | Reviewed by curator |
| Biological Process | GO:0009252 | peptidoglycan biosynthetic process | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
25495032 | Reviewed by curator |
| Biological Process | GO:0006508 | proteolysis |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR30023
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004185 | serine-type carboxypeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR30023
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Peptideglycan biosynthesis |
ECO:0000037
not_recorded |
|||
| KEGG | pae00550 | Peptidoglycan biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| FunFam | G3DSA:3.50.80.20:FF:000002 | D-alanyl-D-alanine carboxypeptidase dacB | - | - | 96 | 163 | 2.6E-47 |
| PRINTS | PR00922 | D-Ala-D-Ala carboxypeptidase 3 (S13) family signature | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 295 | 317 | 2.2E-22 |
| Gene3D | G3DSA:3.40.710.10 | - | IPR012338 | Beta-lactamase/transpeptidase-like | 280 | 468 | 9.7E-55 |
| PRINTS | PR00922 | D-Ala-D-Ala carboxypeptidase 3 (S13) family signature | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 432 | 445 | 2.2E-22 |
| PRINTS | PR00922 | D-Ala-D-Ala carboxypeptidase 3 (S13) family signature | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 68 | 87 | 2.2E-22 |
| PANTHER | PTHR30023 | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 7 | 474 | 0.0 |
| NCBIfam | TIGR00666 | JCVI: D-alanyl-D-alanine carboxypeptidase/D-alanyl-D-alanine-endopeptidase | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 62 | 454 | 5.3E-65 |
| Pfam | PF02113 | D-Ala-D-Ala carboxypeptidase 3 (S13) family | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 30 | 454 | 1.5E-86 |
| SUPERFAMILY | SSF56601 | beta-lactamase/transpeptidase-like | IPR012338 | Beta-lactamase/transpeptidase-like | 27 | 469 | 9.4E-92 |
| Gene3D | G3DSA:3.50.80.20 | - | - | - | 92 | 163 | 7.3E-22 |
| PRINTS | PR00922 | D-Ala-D-Ala carboxypeptidase 3 (S13) family signature | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 355 | 369 | 2.2E-22 |
| PRINTS | PR00922 | D-Ala-D-Ala carboxypeptidase 3 (S13) family signature | IPR000667 | Peptidase S13, D-Ala-D-Ala carboxypeptidase C | 370 | 384 | 2.2E-22 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.