In order to generate a more inclusive dataset of Pseudomonas genes mapped to putative in-paralogs and putative orthologs in other Pseudomonas species/strains, we developed a Pseudomonas Orthologous Groups classification system.
To generate ortholog groups, pair-wise DIAMOND searches were run on all genomes in the database to find reciprocal best hits (RBHs) for each gene. These analyses often resulted in multiple candidate genes for RBH status, which were narrowed down by examining the similarity between the query's flanking genes and the hit's flanking genes. If two candidate genes were directly adjacent, they where both accepted as RBHs that involve putative in-parology.
Pairwise intra-genome DIAMOND searches were also performed to acquire in-paralog information (i.e. gene duplications occurring after species divergence). If two genes in one genome were reciprocally more similar to each other than to any gene in the other genomes, the two genes were designated putative in-paralogs. Ortholog groups are built by starting with a seed gene and then adding all genes to which there is a RBH or in-paralog relationship.
Every new gene added to an ortholog group was then treated as a seed gene and the addition process was repeated until all qualifying genes had been added. The result was the development of orthologous groups, specifically generated for Pseudomonas species genomes, which can be used to sort search results.
Pseudomonas Ortholog Group POG000081
| Strain | Locus Tag | Description | Same-Strain Members | Fragment ? | |
|---|---|---|---|---|---|
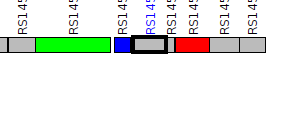
| Pseudomonas syringae UB0390 | IV03_RS14575 |
ImpA, N-terminal
|
1 member |
|
|
| Pseudomonas taeanensis MS-3 - Assembly GCF_000498575.2 | TMS3_RS0118065 |
type VI secretion protein ImpA
|
1 member |

|
|
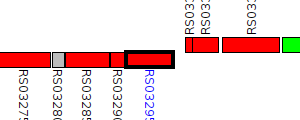
| Pseudomonas taetrolens DSM 21104 | TU78_RS03295 |
type VI secretion protein ImpA
|
1 member |
|
|
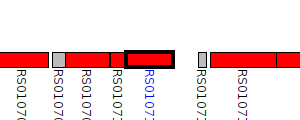
| Pseudomonas taiwanensis DSM 21245 | H620_RS0107105 |
type VI secretion protein ImpA
|
1 member |
|
|
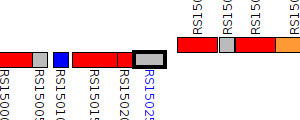
| Pseudomonas trivialis IHBB745 | AA957_RS15025 |
type VI secretion protein ImpA
|
2 same-strain members: AA957_RS15025 AA957_RS27320 |
|
|
| Pseudomonas trivialis IHBB745 | AA957_RS27320 |
type VI secretion protein ImpA
|
2 same-strain members: AA957_RS15025 AA957_RS27320 |

|
|
| Pseudomonas tuomuerensis JCM 14085 | PT85_RS12190 |
hypothetical protein
|
1 member |

|
|
| Pseudomonas umsongensis 20MFCvi1.1 | D470_RS0113790 |
type VI secretion protein ImpA
|
3 same-strain members: D470_RS0113790 D470_RS0113860 D470_RS0126045 |
|
|
| Pseudomonas umsongensis 20MFCvi1.1 | D470_RS0113860 |
type VI secretion protein ImpA
|
3 same-strain members: D470_RS0113790 D470_RS0113860 D470_RS0126045 |
|
|
| Pseudomonas umsongensis 20MFCvi1.1 | D470_RS0126045 |
type VI secretion protein
|
3 same-strain members: D470_RS0113790 D470_RS0113860 D470_RS0126045 |
|
|
| Pseudomonas umsongensis UNC430CL58Col | N519_RS0120505 |
type VI secretion protein
|
3 same-strain members: N519_RS0105210 N519_RS0105310 N519_RS0120505 |

|
|
| Pseudomonas umsongensis UNC430CL58Col | N519_RS0105210 |
type VI secretion protein ImpA
|
3 same-strain members: N519_RS0105210 N519_RS0105310 N519_RS0120505 |
|
|
| Pseudomonas umsongensis UNC430CL58Col | N519_RS0105310 |
type VI secretion protein ImpA
|
3 same-strain members: N519_RS0105210 N519_RS0105310 N519_RS0120505 |
|
|
| Pseudomonas veronii 1YdBTEX2 | H736_RS0130240 |
type VI secretion protein ImpA
|
1 member |
|
|
| Pseudomonas veronii R4 | SU91_RS01275 |
type VI secretion protein ImpA
|
1 member |
|
|
| Pseudomonas viridiflava LMCA8 | RT94_RS06390 |
type VI secretion protein ImpA
|
2 same-strain members: RT94_RS06390 RT94_RS06510 |
|
|
| Pseudomonas viridiflava LMCA8 | RT94_RS06510 |
ImpA-like protein
|
2 same-strain members: RT94_RS06390 RT94_RS06510 |
|
|
| Pseudomonas weihenstephanensis DSM 29166 | TU86_RS19230 |
type VI secretion protein ImpA
|
1 member |
|