Pseudomonas aeruginosa PAO1, PA4501 (opdP)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
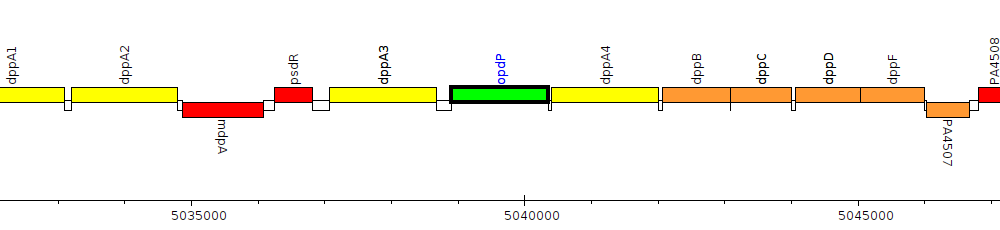
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA4501
|
| Name |
opdP
Synonym: opdP |
| Replicon | chromosome |
| Genomic location | 5038901 - 5040355 (+ strand) |
| Transposon Mutants | 3 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_253191.1 |
| GI | 15599697 |
| Affymetrix | PA4501_at |
| Entrez | 881095 |
| GenBank | AAG07889.1 |
| NCBI Locus Tag | PA4501 |
| protein_id(GenBank) | gb|AAG07889.1|AE004864_1|gnl|PseudoCAP|PA4501 |
| TIGR | NTL03PA04502 |
| UniProtKB Acc | Q9HVS0 |
| UniProtKB ID | Q9HVS0_PSEAE |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
OpdP
|
| Synonyms |
Glycine-glutamate dipeptide porin OpdP OccD3 |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -7.35 |
| Kyte-Doolittle Hydrophobicity Value | -0.479 |
| Molecular Weight (kDa) | 53.0 |
| Isoelectric Point (pI) | 5.80 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 3SYB | MEMBRANE PROTEIN | 07/16/11 | Crystal structure of Pseudomonas aeruginosa OccD3 (OpdP) | Pseudomonas aeruginosa | 2.7 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 16 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG004088 (487 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA4501
Search term: opdP
Search term: OpdP
|
Human Homologs
References
|
Role of the novel OprD family of porins in nutrient uptake in Pseudomonas aeruginosa.
Tamber S, Ochs MM, Hancock RE
J Bacteriol 2006 Jan;188(1):45-54
PubMed ID: 16352820
|
|
Involvement of two related porins, OprD and OpdP, in the uptake of arginine by Pseudomonas aeruginosa.
Tamber S, Hancock RE
FEMS Microbiol Lett 2006 Jul;260(1):23-9
PubMed ID: 16790014
|
|
Responses of carbapenemase-producing and non-producing carbapenem-resistant Pseudomonas aeruginosa strains to meropenem revealed by quantitative tandem mass spectrometry proteomics.
Salvà-Serra F, Jaén-Luchoro D, Marathe NP, Adlerberth I, Moore ERB, Karlsson R
Front Microbiol 2022;13:1089140
PubMed ID: 36845973
|
|
Toward the rational design of carbapenem uptake in Pseudomonas aeruginosa.
Isabella VM, Campbell AJ, Manchester J, Sylvester M, Nayar AS, Ferguson KE, Tommasi R, Miller AA
Chem Biol 2015 Apr 23;22(4):535-547
PubMed ID: 25910245
|
|
Understanding Carbapenem Translocation through OccD3 (OpdP) of Pseudomonas aeruginosa.
Soundararajan G, Bhamidimarri SP, Winterhalter M
ACS Chem Biol 2017 Jun 16;12(6):1656-1664
PubMed ID: 28440622
|
|
High-level resistance to meropenem in clinical isolates of Pseudomonas aeruginosa in the absence of carbapenemases: role of active efflux and porin alterations.
Chalhoub H, Sáenz Y, Rodriguez-Villalobos H, Denis O, Kahl BC, Tulkens PM, Van Bambeke F
Int J Antimicrob Agents 2016 Dec;48(6):740-743
PubMed ID: 28128097
|