Pseudomonas aeruginosa CF614, Q093_02808
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
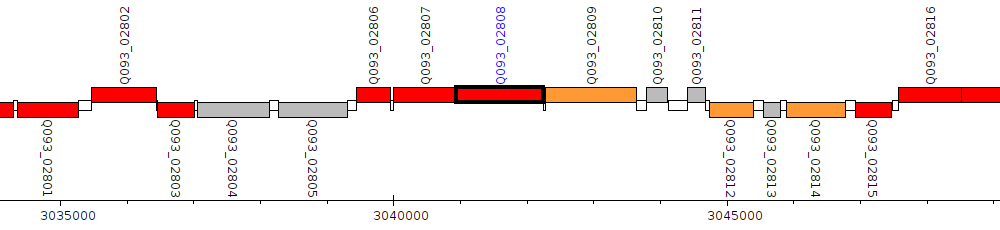
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa CF614
GCF_000480355.1|latest |
| Locus Tag |
Q093_02808
|
| Name |
|
| Replicon | scaffold_1 |
| Genomic location | 3040940 - 3042244 (+ strand) |
Cross-References
| RefSeq | WP_003114643.1 |
| GI | 489205599 |
| NCBI Locus Tag | Q093_02808 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
16S rRNA methyltransferase
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | 5.15 |
| Kyte-Doolittle Hydrophobicity Value | -0.185 |
| Molecular Weight (kDa) | 47.2 |
| Isoelectric Point (pI) | 8.16 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
AlphaFold 2 Protein Structure Predictions
Protein structure predictions using a neural network model developed by DeepMind. If a UniProtKB accession is associated with this protein, a search link will be provided below.
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000017 (536 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: Q093_02808
Search term: 16S rRNA methyltransferase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
NOP2 nucleolar protein [Source:HGNC Symbol;Acc:HGNC:7867]
E-value:
4.8e-27
Percent Identity:
29.0
|
|
Ensembl
110, assembly
GRCh38.p14
|
NOP2 nucleolar protein [Source:HGNC Symbol;Acc:HGNC:7867]
E-value:
4.8e-27
Percent Identity:
29.0
|
|
Ensembl
110, assembly
GRCh38.p14
|
NOP2 nucleolar protein [Source:HGNC Symbol;Acc:HGNC:7867]
E-value:
4.8e-27
Percent Identity:
29.0
|
|
Ensembl
110, assembly
GRCh38.p14
|
NOP2 nucleolar protein [Source:HGNC Symbol;Acc:HGNC:7867]
E-value:
4.8e-27
Percent Identity:
29.0
|
|
Ensembl
110, assembly
GRCh38.p14
|
NOP2 nucleolar protein [Source:HGNC Symbol;Acc:HGNC:7867]
E-value:
4.8e-27
Percent Identity:
29.0
|
|
Ensembl
110, assembly
GRCh38.p14
|
NOP2 nucleolar protein [Source:HGNC Symbol;Acc:HGNC:7867]
E-value:
4.8e-27
Percent Identity:
29.0
|
References
No references are associated with this feature.